
Current status
Open to accept tissue sections and cells from various diseases
Spatial Biology powered by MIST
We offer innovative, individualized solutions through single-cell spatial proteomic assays to optimize your research outcomes. Collaboration benefits each other.



Access our comprehensive set of protocols to streamline your spatial proteomic workflows. We ensure precision, robustness and efficiency in every step.
A comprehensive, open toolkit is provided to analyze MIST array data. It streamlines the process from cell segmentation all the way to typical bioinformatics
Antibody panel
Built upon our MIST-Linker technology that enables conjugation of antibodies with cleavable oligonucleotides, most antibodies can be adopted for MIST spatial proteomics.
Sample requirement
Cultured cells:
- Cell lines: Adherent cells cultured in a 12 or 6 well plate, with confluency 30 - 80%.
- Primary cells: same above. Must ensure monolayer. Do not form clumps or 3D structure.
- Other culture substrates: Adhesive slides that provides excellent cell adhesion.
- Suspension cells need to be fixed on a flat, transparent surface, or in single-cell microwells.
Tissue sections:
- Sectioning at 3 - 5 μm thick. No more than 3 sections / glass slide.
- FFPE and fresh frozen sample sections are acceptable.
- Adhesive slides are preferred that provides excellent tissue adhesion.
- Lay completely flat, no wrinkles, no bubbles, uniform thickness on glass slide.
- Only 3mm x 3mm or 5mm x 5mm sub-region detected for large samples.
Targets and antibodies
-
16 - 24 targets per sample.
-
Antibodies from various vendors are acceptable.
-
Minimally 0.5 μg antibody for each sample.
-
Antibodies with known amount preferred; but unknown ones acceptable.
-
Host species of antibodies are mouse (IgG1, IgG2) and rabbit.
Procedure & Protocols
1. Typical immunostaining using antibodies tagged with oligos through MIST-Linker;
2. Overlay of MIST array with tissue section or attached cells;
3. UV light exposure of assembly;
4. Microscopy scanning of fluorescence. DAPI, Cy3 and Cy5 filters are required.
Requirement:
Fluorescence microscope with motorized stage, or fluorescence slide scanner;
Common UV lamp;
Reagents, pipettors, etc.
The procedure can be downloaded here:
Data analysis
MIST-Explorer is a python-based, open-source program integrates all tools to streamline data analysis for spatial MIST results.

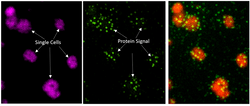
Examples
Here is a set of data from kidney biopsy. Download and use View tab of MIST-Explorer to explore the dataset.
Stay tuned. Will be also visualized by Minerva online.
 |  |  |
|---|---|---|
 |  |  |
 |  |  |
 |  |
Charge
We don't charge any if the sample number is small and if we have available materials and reagents for your samples.
Stay tuned.